WRITE (iocmcf,
& '("MCFGEN Date:", i8, 5x, "Library type:", i3)')
& i_date, nclibtyp
Here, the date is in the format such that 12 January 1995 is
written as 950112. The code for the library type is
-
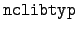 = 0
= 0 - means group-averaged data.
-
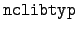 = 1
= 1 - means grid-based data.
WRITE(iocmcf,'("Source Library Id Word:", i12,
& 5x, "Last Changed:", 2x, i6)')
& libid, libdate
Here, libid is meant to be the library identifier packed in
some format, but it is not clear what the components are. The
convention given on page 16 of reference [3] is only followed
for incident photons, because all of the other `mcf.asc'
files have
WRITE(iocmcf,'(i6, 12x, i3, 3x, i3,44x,"*")')
& nnza, negi, numcs
Here, the meaning of the entries is:
- nnza:
- The atomic number and mass number of the
target, formatted as
1000*Z + A. There are some special
conventions. A value of A = 0 denotes the natural element,
so that
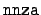 = 8000 is natural oxygen. The value
= 8000 is natural oxygen. The value
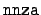 = 99120 denotes a short-lived fission fragment,
and the value
= 99120 denotes a short-lived fission fragment,
and the value
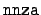 = 999999 is a flag marking the end
of the file.
= 999999 is a flag marking the end
of the file.
- negi:
- The number of energy groups.
- numcs:
- The number of reactions (C-values) for
this target.
- *:
- The final "*" on the line is a flag
showing that we are starting data for a new target.
WRITE(iocmcf,'(1p, i2,4e11.4,1x,i6)')
& iyi, halflife, elvfd, tmpfd, atm, rpdate
The meaning of the entries is:
- iyi:
- The identifier for the incident particle.
- halflife:
- The halflife of the target. A stable target is given a halflife of 1050.
- elvfd:
- The excitation level of the target.
- tmpfd:
- The temperature of the target.
- atm:
- The atomic weight of the target.
- rpdate:
- The date of the data for this target.
WRITE(iocmcf, '(24i3)')
& (C_clean(c_count), c_count=1, numcs)
- Flag:
- A single line, starting with a flag to identify
the type of data. The most common format for the data-flag
line is,
WRITE(iocmcf, '(i4, i7, 1x, a8)') & flag, number, string - Header:
- There is usually a second line identifying the
reaction and the type and number of data entries.
- Data:
- The actual data.
The type of data is identified by means of the flag. The
dentification is unique in all cases except that
![]() = 12
has two interpretations--its most common usage is as the flag for
energy distributions (
= 12
has two interpretations--its most common usage is as the flag for
energy distributions (
![]() = 4), but it also used for
the angular distribution of gammas. The header line must be used
to distinguish them. A point to be aware of is that for
= 4), but it also used for
the angular distribution of gammas. The header line must be used
to distinguish them. A point to be aware of is that for
![]() = 1 and 2 there is no header line. But these two data
blocks appear only with the first target, and they come
immediately after the list of reactions, before the rest of the
data. We list the types of data blocks ordered in terms of flag because that is what is used by the code mcfbin which
reads the `mcf.asc' file. The user who is accustomed to
working in terms of the data identifier I_number should
consult the following list.
= 1 and 2 there is no header line. But these two data
blocks appear only with the first target, and they come
immediately after the list of reactions, before the rest of the
data. We list the types of data blocks ordered in terms of flag because that is what is used by the code mcfbin which
reads the `mcf.asc' file. The user who is accustomed to
working in terms of the data identifier I_number should
consult the following list.
-
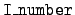 = 0:
= 0: - Cross sections, data with
 = 4, 5, and 6 are given, in that order. For the first
target, however, this data is preceeded by the lists of energy
group boundaries (
= 4, 5, and 6 are given, in that order. For the first
target, however, this data is preceeded by the lists of energy
group boundaries (
 = 1) and target temperatures
(
= 1) and target temperatures
(
 = 2).
= 2).
-
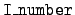 = 1:
= 1: - Angular distributions,
 = 10 and sometimes 12. When
= 10 and sometimes 12. When
 = 12
data is present, it comes first.
= 12
data is present, it comes first.
-
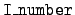 = 3:
= 3: - Correlated energy-angle
distributions,
 = 11.
= 11.
-
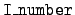 = 4:
= 4: - Energy distributions,
 = 12.
= 12.
-
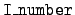 = 7:
= 7: - Fission neutron
multiplicities,
 = 3.
= 3.
-
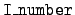 = 9:
= 9: - Photon multiplicities,
 = 9.
= 9.
-
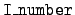 = 10:
= 10: - Energy deposition,
 = 7.
= 7.
-
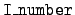 = 12:
= 12: - Average available energy,
 = 6.
= 6.
-
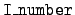 = 941:
= 941: - Coherent scattering
form factors,
 = 14.
= 14.
-
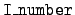 = 942:
= 942: - Incoherent scattering
form factors,
 = 15.
= 15.
The content of the data blocks as ordered by flag is as follows.
-
 = 1:
= 1: - This block of data is energy group
boundaries, number is the number of groups, and string is "gp bds". There is no header, and the energy
group boundaries follow immediately, written with the format,
WRITE(iocmcf, '(1p, 6e11.4)') (ebound(j), j=1, nebi)
-
 = 2:
= 2: - This block of data is target
temperatures, number is the number of temperatures, and
string is "temp.". There must be only one
temperature because Doppler broadening is not now done in mcfgen. That temperature is given on the next line in the
format,
WRITE(iocmcf, '(1p, 6e11.4)') & (temture(j), j=1, numtemp) -
 = 3:
= 3: - This block of data is multiplicity of
fission neutrons. The format of the flag line is nonstandard,
WRITE(iocmcf, '(i4, i2, 1x, a16)') & flag, number, stringFor prompt neutrons we have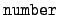 = 0 and the label
string is "nubar, prompt". For delayed neutrons
we have the identifier
= 0 and the label
string is "nubar, prompt". For delayed neutrons
we have the identifier
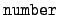 = 1 and string is
"nubar, delayed". The flag line is followed by a header
of the form,
= 1 and string is
"nubar, delayed". The flag line is followed by a header
of the form,
WRITE(iocmcf, '(1p, 5i5, 5x, 3e12.5)') & c, I_number, s, igsrt, igend, q0, x1The meaning of these entries is:- c:
- The C reaction identifier (
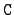 = 15 for fission).
= 15 for fission).
- I_number:
- For
 = 3 the data identifier I_number is 7.
= 3 the data identifier I_number is 7.
- s:
- The S reaction identifier. It
determines the meaning of x1.
- igsrt:
- Identify the first nonzero cross
section. (This amounts to an energy threshold.)
- igend:
- Identify the highest nonzero cross
section. (In effect, this is a maximum energy.)
- q0:
- The energy of the reaction.
- x1:
- The first extra information, depending on
the value of s.
WRITE(iocmcf, '(1p, 6e11.4)') & (array(j), j=igsrt, igend) -
 = 4:
= 4: - This block of data is flux-weighted
group-averaged cross sections, number is the date for
the data, and string is "gp x-sec". The flag
line is followed by a header of the form,
WRITE(iocmcf, '(1p, 6i5, 3e12.5)') & c, I_number, s, igsrt, igend, & kin_type, q0, x1, temptureThe differences from the header for = 3 are
= 3 are
- I_number:
- The data identifier is 0.
- kin_type:
- The kinematics type for this reaction.
- tempture:
- The temperature of the target.
The cross section data follows,
and it is written in the following format.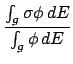
WRITE(iocmcf, '(1p, 6e11.4)') & (gpsig0(j), j=igsrt, igend)The cross section data is followed immediately in the file (without an identification flag) by a line containing the number of particles produced by this reaction, including the residual nucleus.
WRITE(iocmcf, '(i2)') parts_produced(1, nrc)Then come the secondary particles and their multiplicities as pairs (yo, multiplicity). The residual nucleus is the last particle, unless it is light enough to be included with the other secondary particles.WRITE(iocmcf, '(15i6)') (parts_produced(j, nrc), & j=2, parts_produced(1, nrc)*2 + 1) -
 = 5:
= 5: - This block of data is the calculated
energy available, and it always follows the block for
 = 4. The flag line is nonstandard in that number
is not present. The string in the flag line is
"en avail". The header line from
= 4. The flag line is nonstandard in that number
is not present. The string in the flag line is
"en avail". The header line from
 = 4 is
printed again. (We again have
= 4 is
printed again. (We again have
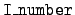 = 0.)
The available energies per group
are then given with the format,
= 0.)
The available energies per group
are then given with the format,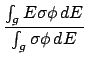
WRITE(iocmcf, '(1p, 6e11.4)') & (av_en_avail(j), j=igsrt, igend) -
 = 6:
= 6: - If this block of data is present, it
appears immediately after the block for
 = 5. It
contains the energy available as given in the Production Cross
Section library file. In the flag line number is not
present, and string is "Q-bar". The header line
from
= 5. It
contains the energy available as given in the Production Cross
Section library file. In the flag line number is not
present, and string is "Q-bar". The header line
from
 = 4 is printed again. (We again have
= 4 is printed again. (We again have
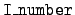 = 0, but now
= 0, but now
 = 5.) The available
energies per group are then given with the format,
= 5.) The available
energies per group are then given with the format,
WRITE(iocmcf, '(1p, 6e11.4)') & (qbargp(j), j=igsrt, igend) -
 = 7:
= 7: - This block of data is energy carried by
a secondary particle, number identifies the first of the
list of secondary particles, and string is "edeptoyo". The flag line is followed by the same header as
for
 = 3. The only difference is that the data
identifier I_number is now 10. The data is
and is given in the format,
= 3. The only difference is that the data
identifier I_number is now 10. The data is
and is given in the format,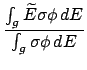
WRITE(iocmcf, '(1p, 6e11.4)') & (e10barg(io), io = igsrt, igend) -
 = 9:
= 9: - This block of data is group-averaged
photon multiplicity, number identifies the first of the
list of secondary particles, and we have "multipl." for
string. The flag line is followed by the same header as
for
 = 3. The only difference is that the data
identifier I_number is now 9. The data
is in the format,
= 3. The only difference is that the data
identifier I_number is now 9. The data
is in the format,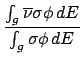
WRITE(iocmcf, '(1p, 6e11.4)') & (array(j), j=igsrt, igend) -
 = 10:
= 10: - This block of data gives equiprobable
angular bins. The number identifies the first of the
list of secondary particles, and we have "ang(i=1)" for
string. The flag line is followed by the header,
WRITE(iocmcf, '(1p, 4i5, 5x, i5, 3e12.5)') & c, I_number, s, num_engy_in, neqpb, q0, x1For this value of the flag the data identifier I_number is 1. The unusual item in the header is neqpb, the number of equiprobable angular bin edges. (The number of bins is one less.) The first data entries are the incident energies in the format,WRITE(iocmcf, '(1p, 6e11.4)') & (energy_in(k), k=1, num_engy_in)The equiprobable angular bin boundaries are concatenated into a single long array, with a block for each incident energy. The entries are printed in the format,WRITE(iocmcf, '(1p, 6e13.6)') & (cpdi2(k), k=1, neqpb*num_engy_in) -
 = 11:
= 11: - This block of data gives correlated
bins which are equally probable jointly with respect to angle
and energy. The number in the flag line identifies the
first of the list of secondary particles, and string is
"ang(i=3)". The flag line is followed by the header,
WRITE(iocmcf, '(1p, 6i5, 3e12.5)') & c, I_number, s, num_engy_in, & nppbe, neqint, q0, x1Here, the value of I_number is 3. The number of equiprobable angular bin edges is
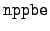 , and the number
of equiprobable energy bin edges is
, and the number
of equiprobable energy bin edges is
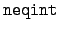 . Just as
for
. Just as
for
 = 10 (equiprobable energy distributions), the
block starts with the incident energies in the format,
= 10 (equiprobable energy distributions), the
block starts with the incident energies in the format,
WRITE(iocmcf, '(1p, 6e11.4)') & (energy_in(k), k=1, num_engy_in)The joint energy-angle distributions are printed in sections. Each section contains, first, an equiprobable angular bin boundary point
WRITE(iocmcf, '(1p, 6e11.4)') & I1_equi_cos(k, engy_index)There follows the equiprobable energy bin boundaries for this cosine,WRITE(iocmcf, '(1p, 6e13.6)') & (equi_engy(j), j=1, neqint) -
 = 12:
= 12: - This flag has two possible meanings,
and they ought to be split. In both cases number
identifies the first of the list of secondary particles, and
string is "e spec". The two meanings of the flag
are distinguished by different header lines.
-
 = 12, Energy distributions:
= 12, Energy distributions: - If
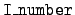 = 4 the flag 12 denotes equiprobable energy
distributions. The header is,
= 4 the flag 12 denotes equiprobable energy
distributions. The header is,
WRITE(iocmcf,'(1p, 4i5, 5x, i5, 3e12.5)') & c, I_number, s, num_engy_in, & neqpb, q0, x1This is standard header information except for neqpb, the number of equiprobable energy bin edges. (The number of bins is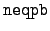 - 1.) The data is in two
sets. First, we have the incident energies,
- 1.) The data is in two
sets. First, we have the incident energies,
WRITE(iocmcf, '(1p, 6e11.4)') & (energy_in(k), k=1 ,num_engy_in)The equidistributed energy deposition bin boundaries for the various incident energies are concatenated into a single array, and this array is printed in the format,WRITE(iocmcf, '(1p, 6e13.6)') & (cpdi2(k), k=1, neqpb*num_engy_in) -
 = 12, Gamma production:
= 12, Gamma production: - If
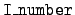 = 1 and
= 1 and
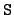 = 3 (so that the data gives
angular distribution for gamma production), this block
appears before the angular distribution data (which has
= 3 (so that the data gives
angular distribution for gamma production), this block
appears before the angular distribution data (which has
 = 10). There is a special header,
= 10). There is a special header,
WRITE(iocmcf, '(1p, 5i5, 5x, 3e12.5)') & c, I_number, s, 2, 2, q0, x1In this case x1 is the energy of the ejected gamma. This line is followed by the energy rangeWRITE(iocmcf, '(1p, 6e11.4)') emin, emaxand by the energy of the ejected gamma, four more times!WRITE(iocmcf, '(1p, 4e13.6)') (x1, i0 = 1, 4)Note that this is the only situation in which the values of emin and emax get printed, even though they are present with all of the data in the `library' file.
-
-
 = 14:
= 14: - This block of data gives coherent
scattering form factors. In the flag line number
identifies the first of the list of secondary particles, and
string is "coh.scat". The flag line is followed
by the header,
WRITE(iocmcf, '(1p, 4i5, 2e12.5)') & c, I_number, s, ndata, q0, x1Here, the value of I_number is 941, and ndata is the length of the data. The scattering form factors are given in the format,WRITE(iocmcf, '(1p, 6e11.4)') & (scats(j), j=1, ndata) -
 = 15:
= 15: - This block of data gives incoherent
scattering form factors. In the flag line number
identifies the first of the list of secondary particles, and
string is "inc.scat". The header and the data
block are the same as for
 = 14. The only
difference is that I_number is 942.
= 14. The only
difference is that I_number is 942.
-
 = 99:
= 99: - There is no more data for this target.